In the world of plants, Pseudomonas syringae is a notorious troublemaker. This soil-dwelling bacterium is responsible for devastating blights, leaf spots, and frost damage across agricultural fields worldwide. But beneath its destructive reputation lies a remarkable chemical factory—a hidden arsenal of molecules that could help scientists fight not just plant disease, but potentially human infections and fungal threats as well.
A new study, published in Angewandte Chemie International Edition, reveals that P. syringae isn’t just a threat—it’s also an unexpected source of inspiration. By decoding the bacterial genome and analyzing its chemical potential, an interdisciplinary team of researchers has uncovered two entirely new families of bioactive compounds. These microscopic molecules, called syrilipamides and secimides, have powerful effects on microorganisms, killing fungi and even amoebae with startling precision.
Far from being just another pathogen, P. syringae now emerges as a biochemical innovator—one that may someday shape the future of antibiotics, plant protection, and biotechnology.
A Hidden Arsenal in the Genome
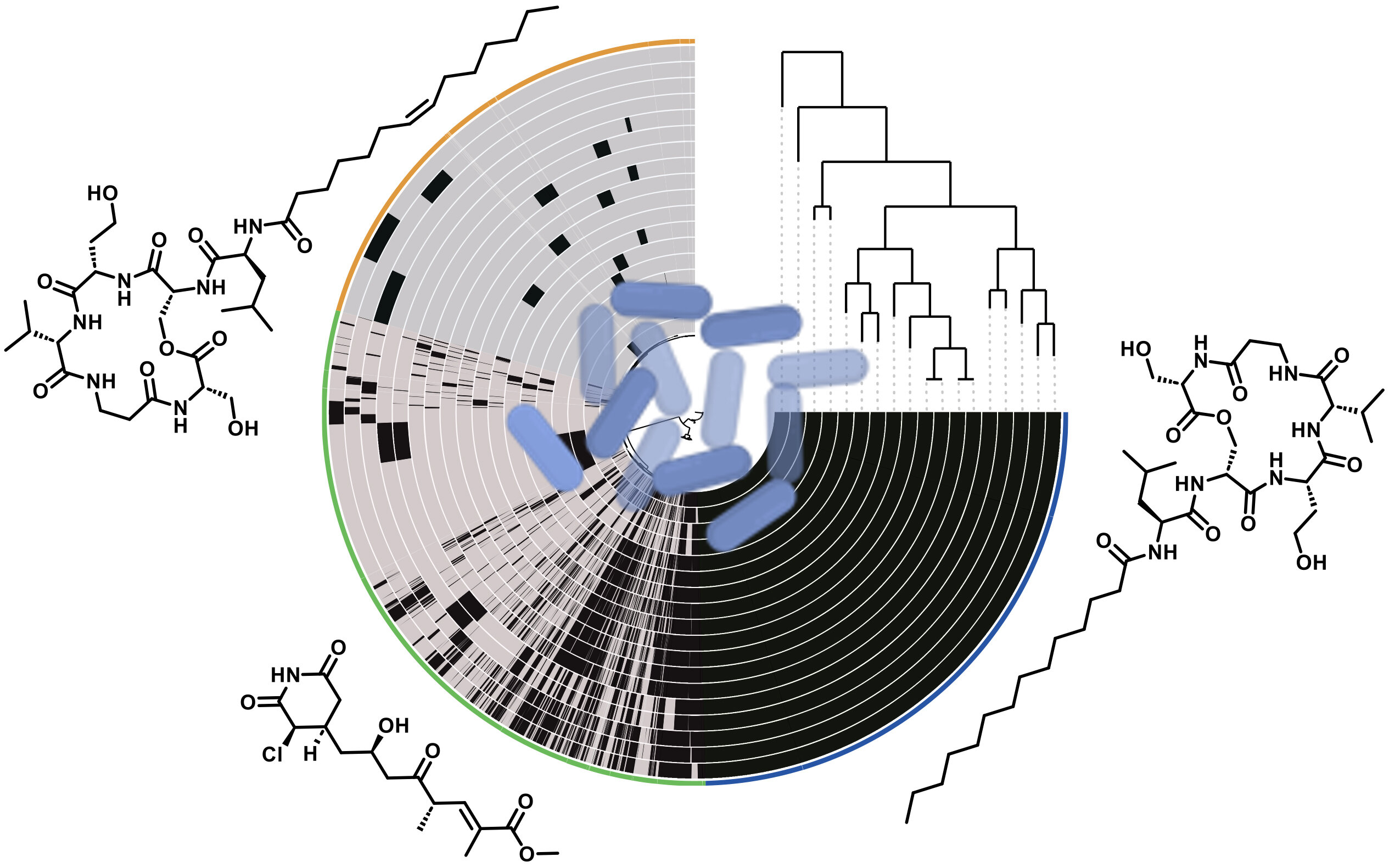
The research team, drawn from the Leibniz Institute for Natural Product Research and Infection Biology (Leibniz-HKI) and the Friedrich Schiller University Jena, took a deep dive into the genetic blueprint of P. syringae. They selected 18 representative strains of the bacterium and scanned their DNA using cutting-edge bioinformatics tools, searching for the microbial equivalent of buried treasure: biosynthetic gene clusters.
These clusters are groups of genes that work together like a factory assembly line, encoding enzymes that build complex natural products. What the team found was staggering—231 distinct biosynthetic gene clusters, a number that suggests P. syringae is far more chemically diverse than previously imagined.
Many of these clusters code for nonribosomal peptide synthetases (NRPS), special enzymes that allow bacteria to create unique peptide-based compounds without using the typical genetic code. These peptides often serve as potent weapons—antibiotics, antifungals, or toxins designed to suppress rival microbes in the wild.
In other words, P. syringae is armed to the teeth.
Discovering New Molecular Families
From this biochemical treasure trove, two new families of small molecules emerged: syrilipamides and secimides.
These compounds are tiny but potent. Initial tests showed that both types are especially toxic to amoebae, microorganisms that often feed on bacteria. This suggests that P. syringae uses them as a line of defense—chemical armor against microbial predators. Even more intriguingly, the secimides were found to be highly active against fungi as well, indicating a dual-purpose effect.
“Both of these natural product families exhibit remarkable selectivity,” said Shuaibing Zhang, the study’s lead author. “They don’t just kill indiscriminately—they seem to be tuned to target very specific threats in the bacterium’s environment.”
Such precision makes these molecules particularly appealing as potential candidates for new antibiotics or antifungal drugs, especially at a time when resistance to existing treatments is a growing global crisis.
A New Enzyme Lights the Way
Beyond the discovery of new molecules, the study also brought an unexpected star to the surface: a previously unknown enzyme called SecA.
This enzyme performs a kind of biochemical magic. It adds chlorine atoms to the syrilipamides and secimides, subtly tweaking their structure—but massively increasing their potency.
“Adding chlorine atoms increases the structural complexity and, in many cases, the biological activity of these compounds,” explained Pierre Stallforth, senior author of the study and head of the research department at Leibniz-HKI. “This modification is a valuable tool in drug discovery.”
In fact, chlorinated natural products are a hot topic in pharmaceutical research. Many widely used medications—ranging from cancer drugs to antibiotics—feature halogen atoms like chlorine. The ability to produce such molecules biologically, using enzymes like SecA, opens up new avenues for synthetic biology and sustainable drug development.
Nature’s Microbial Chess Game
So why does a soil bacterium invest so much energy into making complex molecules?
The answer lies in ecology. In the wild, life is a constant competition. Microbes battle for nutrients, space, and survival. Every molecule that gives P. syringae an edge—by repelling predators, killing competitors, or adapting to new environments—is an evolutionary asset.
“These molecules aren’t random,” said Stallforth. “They’re survival tools. They give the bacterium the ability to thrive in a constantly changing and competitive habitat.”
This dynamic is especially relevant in agriculture. P. syringae is infamous for attacking crops, but its own defense mechanisms might also be used to protect plants from other microbial threats. The very substances that make it a problem could be turned into solutions—if we can harness them properly.
From Plant Pathogen to Biotech Hero?
While it’s too early to say if syrilipamides or secimides will become commercial products, their discovery expands our understanding of microbial chemistry—and highlights the enormous potential hidden in the soil beneath our feet.
“We started this research to better understand P. syringae as an agricultural pathogen,” said Zhang. “What we found was a complex microbial strategist, equipped with a biochemical toolkit that’s still largely unexplored.”
In the long term, these findings could help create new antifungal treatments, plant-safe biopesticides, or even novel antibiotics. But more immediately, they underscore the power of looking at nature not just as a battlefield, but as a library—a living archive of molecular possibilities shaped by millions of years of evolution.
Peering Into the Microverse
This work is part of a larger effort within the Jena Cluster of Excellence “Balance of the Microverse,” a research initiative dedicated to understanding the complex relationships between microbes and their environments. In nature, bacteria rarely act alone. They form dynamic communities—microbial consortia—that interact in subtle and often surprising ways.
By studying these interactions, scientists hope to unravel how life really functions at the microscopic level—not just in disease, but in health, agriculture, ecosystems, and even climate science.
The discovery of the syrilipamides and secimides is just one chapter in this unfolding story. But it’s a vivid reminder that even the most unassuming organisms—like a soil bacterium that kills crops—may carry inside them the keys to solving some of our greatest scientific challenges.
Reference: Shuaibing Zhang et al, Pangenome Analysis of the Plant Pathogen Pseudomonas syringae Reveals Unique Natural Products for Niche Adaptation, Angewandte Chemie International Edition (2025). DOI: 10.1002/anie.202503679