In a groundbreaking study blending innovation and perseverance, researchers from Oregon Health & Science University and Oregon State University have pulled back the molecular curtain on one of biology’s hidden highways: a massive protein complex called LPD-3. Published recently in Nature, their work reveals the astonishing structural organization and lipid-ferrying capabilities of LPD-3, shedding light on how cells move essential lipid molecules between membranes — a process crucial to life itself.
This discovery not only fills a major gap in cell biology but also opens promising avenues for understanding diseases like Alkuraya-Kučinskas syndrome, a severe human neurological disorder linked to mutations in the LPD-3 family.
The Challenge of Moving Lipids: Nature’s Transport Problem
Inside every living cell, membranes are the unsung heroes. They guard the cell’s interior, coordinate communication, and manage the flow of nutrients and information. Their strength and function depend largely on lipids — greasy molecules manufactured mainly in the endoplasmic reticulum (ER).
Yet moving lipids poses a tricky conundrum: because they are hydrophobic (water-fearing), they cannot simply drift through the watery cytoplasm. Evolution answered this challenge by creating lipid-transfer proteins that act as molecular shuttles. For decades, scientists knew that small lipid carriers existed, ferrying a few molecules at a time between organelles. But how cells managed large-scale lipid movement remained a mystery.
Emerging evidence pointed to a fascinating solution: bridge-like lipid-transfer proteins (BLTPs) — gigantic molecular structures hypothesized to form direct tunnels between membranes, allowing lipids to glide en masse from one place to another. However, due to their sheer size and complexity, the actual structure and workings of BLTPs had remained stubbornly elusive.
Until now.
Cracking the Structure of LPD-3
The team at Oregon took on the daunting task of mapping the architecture of LPD-3, a prototype BLTP. Their approach combined cutting-edge cryogenic electron microscopy (cryo-EM) with mass spectrometry, focusing on the native LPD-3 complex isolated from genetically engineered strains of the humble nematode Caenorhabditis elegans.
Two custom C. elegans strains were created, each with fluorescent and epitope tags inserted at strategic points on the natural lpd-3 gene. After confirming that these modifications did not impair normal development or stress resistance, researchers embarked on an epic purification effort. It took about 60 million worms to collect enough material for structural and biochemical analysis.
Using fluorescence-based size-exclusion chromatography, they isolated the massive LPD-3 complex and analyzed it by mass spectrometry, identifying peptides spanning the protein’s enormous 4,022 amino acid sequence. This alone was a technical triumph — few protein studies wrangle molecules this large and unwieldy.
Cryo-EM, which allows imaging at near-atomic resolutions by freezing samples and bombarding them with electrons, yielded a 6.2 Ångstrom-resolution map of LPD-3’s full length. Though some regions — particularly the C-terminal half — proved too flexible to resolve fully, the scientists were able to piece together an astonishing picture.
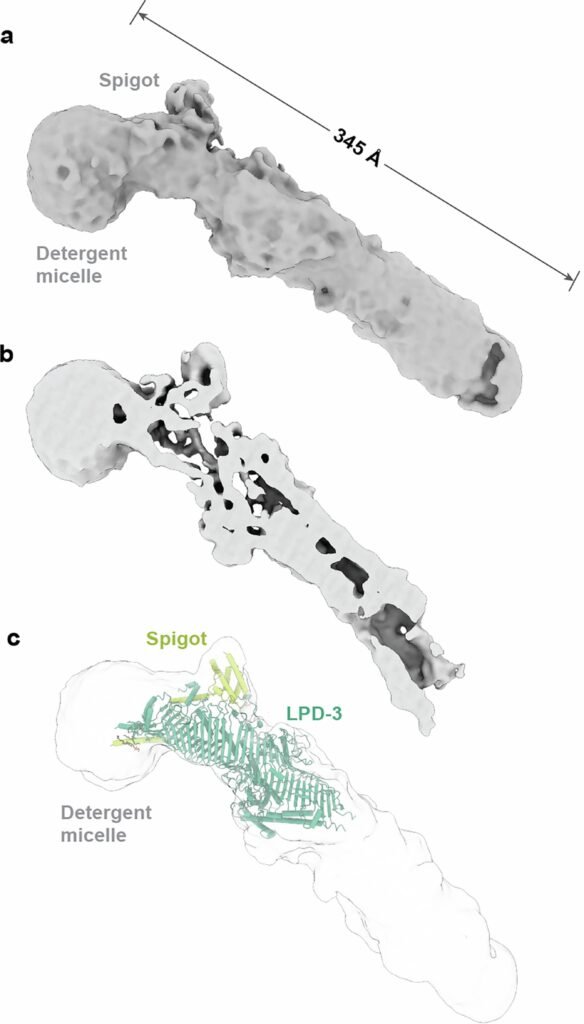
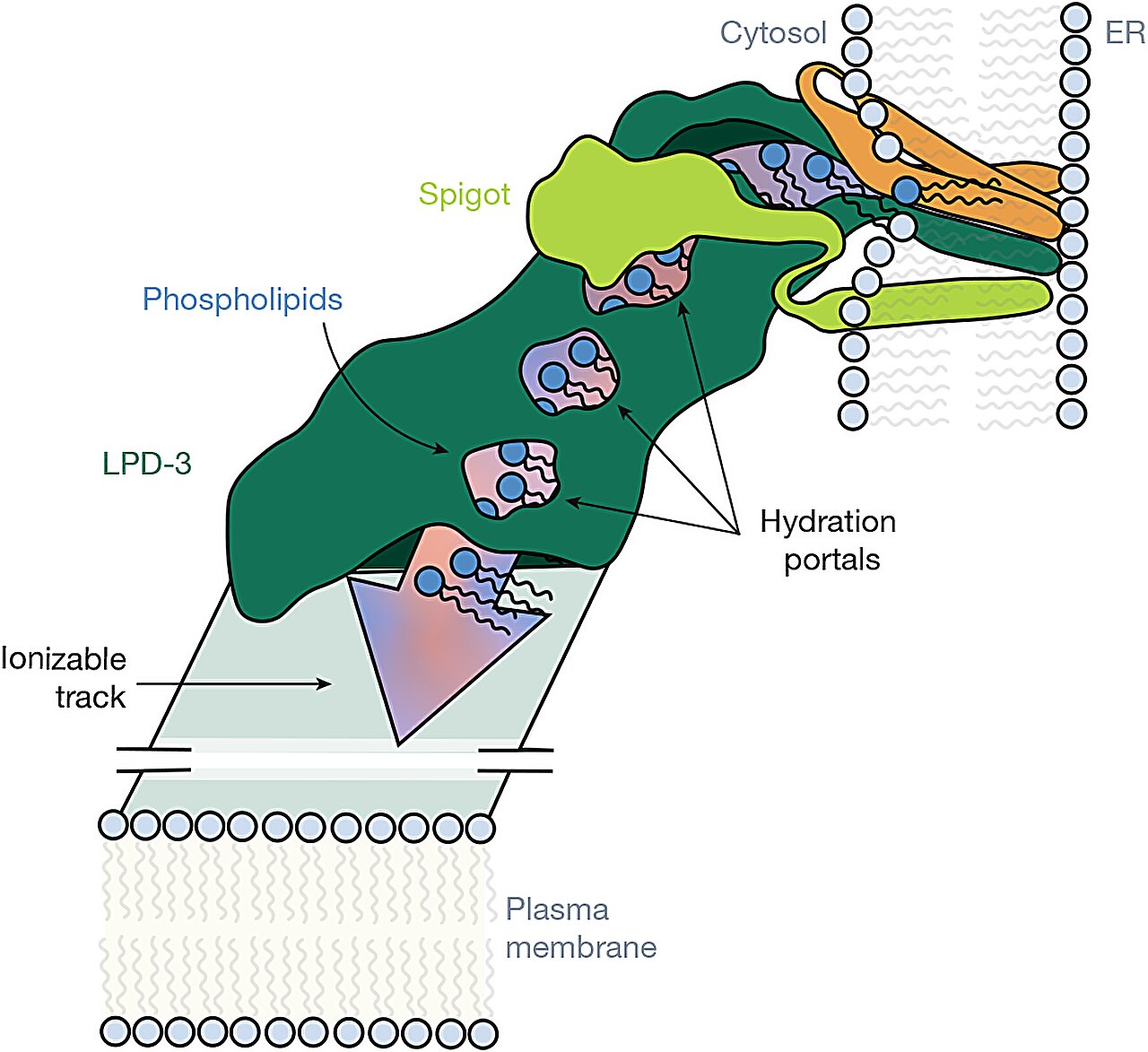
A Tunnel Built for Lipids
LPD-3, it turns out, is not just large. It’s monumental. Measuring roughly 345 Ångstroms in length (about 35 nanometers), it resembles a giant molecular pipeline bridging membranes.
Inside, the structure harbors an elongated, hydrophobic tunnel, cradling 27 lipid molecules lined up like train cars on a track. At the transmembrane region anchoring LPD-3 into the ER membrane, three additional phospholipids were observed nestled inside. The lipids within the tunnel were spaced about 8.4 Ångstroms apart — a spacing eerily similar to the arrangement of lipid molecules in natural bilayers.
The tunnel’s interior isn’t smooth: it is lined by alternating acidic and basic amino acid residues, creating an ionizable track that likely helps organize and shepherd lipid headgroups during transfer. Intriguingly, four hydration portals connect the tunnel to the surrounding cytosol, letting water molecules sneak in and interact with the polar heads of lipids while their greasy tails remain safely embedded.
This structural arrangement suggests a robust, efficient system for bulk lipid transfer, vastly faster and more effective than small shuttle proteins.
More Than Just LPD-3: Auxiliary Players Join the Scene
In a surprise twist, mass spectrometry revealed that LPD-3 doesn’t work alone. Two co-purifying auxiliary proteins were identified.
The first, named Spigot, binds near the N-terminal region of LPD-3 and exhibits strong conservation across species — from worms to humans. The second, dubbed lipid transfer auxiliary protein 2 (LTAP2), also appeared conserved but its precise placement within the LPD-3 structure remains unclear.
Additionally, researchers noticed a third component — a mysterious three-helix transmembrane bundle — whose identity and function are yet to be unraveled.
Functional experiments showed just how vital these auxiliary proteins are. Using RNA interference (RNAi) to knock down the spgt-1 gene encoding Spigot in C. elegans led to a 79.1% reduction in apical actin fluorescence, indicating serious disruptions to membrane structure and cell polarity. Knocking down lpd-3 itself had even more dramatic consequences, slashing actin fluorescence by 91.6%.
These findings were not limited to worms. When Spigot orthologs were silenced in Drosophila (fruit flies), astrocytes — the brain’s support cells — lost their ability to efficiently engulf and clear debris. In HeLa cells (human cells derived from cervical cancer), knocking down the human Spigot ortholog C1orf43 severely impaired the formation of ER–plasma membrane contact sites, crucial hubs for inter-organelle communication.
Taken together, these results established Spigot as a vital, conserved partner in LPD-3’s lipid transport mission.
Implications for Human Health: Alkuraya-Kučinskas Syndrome
Beyond illuminating basic cell biology, this work holds deep relevance for medicine.
Mutations in human BLTP1, the gene encoding LPD-3’s human cousin, are implicated in Alkuraya-Kučinskas syndrome — a devastating condition marked by severe neurological deficits, developmental delays, and growth abnormalities. Understanding the LPD-3 structure offers a critical framework for deciphering how mutations cripple its function and, ultimately, disrupt cellular organization and brain development.
Moreover, by revealing detailed features like hydration portals, ionizable tracks, and lipid stacking, the study sets the stage for future therapeutic strategies — perhaps small molecules or gene therapies designed to stabilize or mimic defective bridge-like lipid transporters.
The Road Ahead: Many Bridges Yet to Be Crossed
This study represents a milestone in cell biology: the first complete glimpse at a bridge-like lipid-transfer protein in its native, functional state. Yet questions abound.
What is the exact role of LTAP2 and the unknown three-helix bundle? How dynamic is lipid transport through the LPD-3 tunnel under different cellular conditions? Can manipulating hydration portals or ionizable residues tweak transport efficiency?
Further, the relationship between LPD-3 structures and other members of the vast BLTP family awaits exploration. Each might feature unique twists — different tunnel dimensions, gating mechanisms, or lipid specificities — tailored to the specific demands of diverse cell types.
The implications ripple far beyond fundamental science. In a world increasingly aware of the delicate ties between cell structure, health, and disease, understanding the infrastructure of lipid transport could offer novel tools to tackle disorders ranging from neurodegeneration to cancer.
Conclusion: The Beauty of Biological Engineering
The unveiling of LPD-3 is a reminder that nature’s engineering rivals our wildest imaginations. Within the tiny confines of a cell, vast, elegant structures orchestrate life’s critical processes with breathtaking precision.
Through painstaking experimentation, Oregon’s scientists have not only decoded a molecular marvel but have also opened a window into the intimate choreography of life — a performance of lipids, proteins, and water molecules dancing along the corridors of the LPD-3 highway.
And in that dance lies the key to future scientific breakthroughs, medical treatments, and perhaps even new ways of understanding the cellular fabric that binds us all.
Reference: Yunsik Kang et al, Structural basis of lipid transfer by a bridge-like lipid-transfer protein, Nature (2025). DOI: 10.1038/s41586-025-08918-y