On a quiet morning in an Australian laboratory, a revolution unfolded—not with the roar of engines or the crackle of rocket fuel, but in the invisible world of proteins.
In mere seconds, a computer spat out a molecule that could destroy one of humanity’s deadliest foes: drug-resistant bacteria. It was a protein that didn’t exist in nature. It had never evolved through eons of survival-of-the-fittest. Instead, it was designed, atom by atom, by artificial intelligence.
And it worked.
This astonishing leap, led by researchers from the University of Melbourne’s Bio21 Institute and Monash Biomedicine Discovery Institute, marks the first time Australian scientists have used AI to generate a fully functional biological protein—a weapon capable of killing antibiotic-resistant superbugs like E. coli.
Their findings, published in Nature Communications, hint at a future where humanity designs new medicines as quickly as typing a prompt into a search bar. A future where AI becomes our co-pilot in the battle against disease, venom, and even cancer.
A Problem Older Than Penicillin
To understand why this matters so much, we have to remember the quiet war raging inside hospitals and communities around the world.
Bacteria are evolving. Once treatable infections are now shrugging off entire shelves of antibiotics. Each year, drug-resistant infections kill more than a million people globally—a grim number that’s projected to skyrocket if we don’t find new solutions.
Traditional drug development is agonizingly slow. Designing a custom-made protein—a molecule precisely shaped to bind to a target, like a microscopic key fitting into a lock—can take decades of lab work, trial and error, and millions of dollars.
But now, something fundamentally new is happening. Computers are learning the language of life itself.
The Machine That Builds Molecules
At the center of this breakthrough are two Australian scientists: Dr. Rhys Grinter and Associate Professor Gavin Knott. Along with a team of brilliant structural biologists and computer scientists, they’ve built Australia’s first AI Protein Design Platform—a digital foundry where proteins can be imagined, sculpted, and tested in silico before they ever touch a petri dish.
Their work builds on tools pioneered by American biochemist David Baker, who last year received the Nobel Prize in Chemistry for cracking the code of protein design.
“Proteins are now being developed as pharmaceuticals, vaccines, nanomaterials and tiny sensors, with many other applications yet to be tested,” said Associate Professor Knott.
The magic of the Australian team’s platform lies in deep learning—a class of AI inspired by how neurons in our brains connect and learn. Fed with mountains of protein structures and sequences, the system has learned the rules of folding, binding, and biological function.
It can generate thousands of entirely new proteins in moments, each tailored for a specific job: killing bacteria, neutralizing snake venom, halting the growth of tumors.
A Protein That Kills Superbugs
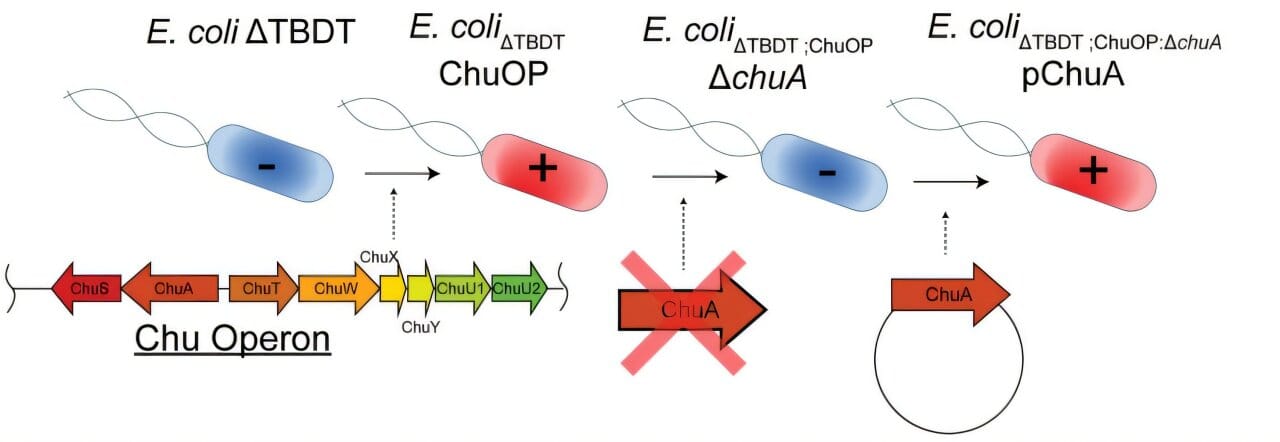
In this latest study, the team set their sights on one of the most formidable enemies in modern medicine: antibiotic-resistant E. coli.
Guided by algorithms, their AI designed proteins that could bind to specific molecules on the surface of these bacteria—like picking the perfect lock to break in and kill.
Ph.D. student Daniel Fox, who carried out much of the experimental work, emphasized the power of freely available AI tools in the process.
“It’s important to democratize protein design so that the whole world has the ability to leverage these tools,” Fox said. “Using these tools and those we are developing in-house, we can engineer proteins to bind a specific target site or ligand, as inhibitors, agonists or antagonists, or engineered enzymes with improved activity and stability.”
Unlike traditional methods, which rely on harvesting proteins from nature and tweaking them, the AI platform designs these molecules from scratch—what scientists call de novo design.
“New methods in deep learning enable efficient de novo design of proteins with specific characteristics and functions, lowering the cost and accelerating the development of novel protein binders and engineered enzymes,” said Dr. Grinter.
Beyond Bacteria: A Universe of Possibilities
While superbugs were the first target, the implications reach far wider. Imagine custom proteins that disarm deadly viruses, block cancer cells from spreading, neutralize toxins in snake bites, or serve as molecular sensors detecting disease at the earliest stages.
These aren’t sci-fi dreams anymore. AI protein design has become a new frontier in biotechnology—an intersection of biology, physics, and machine learning.
New tools like Bindcraft and Chai, developed since Baker’s pioneering work, are already being integrated into the Australian platform. They’re pushing the limits of what’s possible, enabling more precise and sophisticated designs.
Professor John Carroll, Director of the Monash Biomedicine Discovery Institute, called the program a triumph of both science and determination.
“The new AI Protein Design Program brings Australia right up to speed in this exciting new modality for designing novel therapeutics and research tools,” Carroll said. “It is testament to the entrepreneurial spirit of two fabulous young scientists who have worked night and day to build this capability from scratch.”
The Democratization of Molecular Medicine
Perhaps the most thrilling part of this story is that the tools aren’t locked away behind corporate paywalls. The team is committed to open science—sharing software, datasets, and methods so researchers around the world can design their own molecules.
It’s a scientific renaissance unfolding before our eyes. Where once a handful of pharmaceutical giants held the keys to drug discovery, now even small labs can design molecular machines to solve local health crises.
It’s not just about speed or cost—it’s about equity. It’s about a world where life-saving medicines could be designed in weeks rather than decades, for diseases that big pharma ignores because there’s no profit to be made.
A Future Written in Code and Molecules
This is only the beginning. As AI models grow smarter and more powerful, their ability to generate precise, effective, and safe proteins will transform how humanity fights disease.
The researchers behind this Australian breakthrough know the road ahead is still long. Proteins must be tested for safety, efficacy, and stability inside the human body. Regulatory hurdles remain. Evolution always finds ways to fight back.
But for now, there’s a tangible sense of hope—a belief that we’re stepping into a new era where biology and artificial intelligence collaborate to write the next chapter of medicine.
Some revolutions are loud and explosive. Others happen quietly, molecule by molecule, in the glow of a computer screen.
And somewhere in an Australian lab, a machine is dreaming up the molecules that might save us all.
Reference: Inhibiting heme piracy by pathogenic Escherichia coli using de novo-1 designed proteins, Nature Communications (2025). DOI: 10.1038/s41467-025-60612-9 On BioRxiv. DOI: 10.1101/2024.12.05.626953