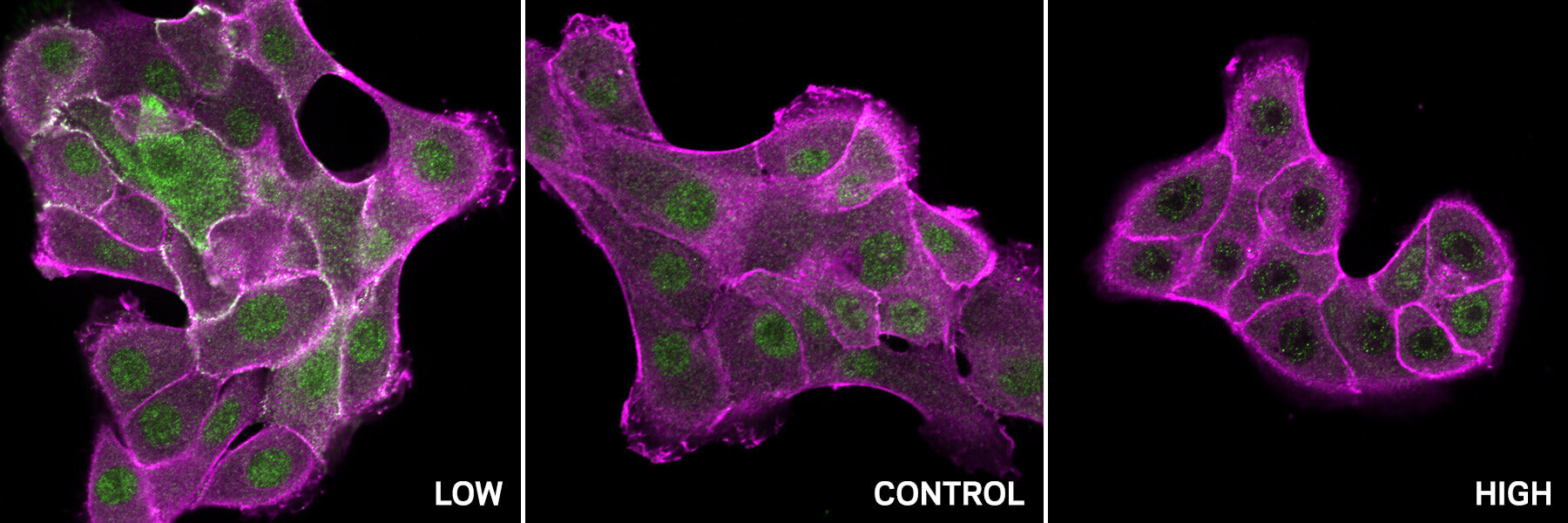
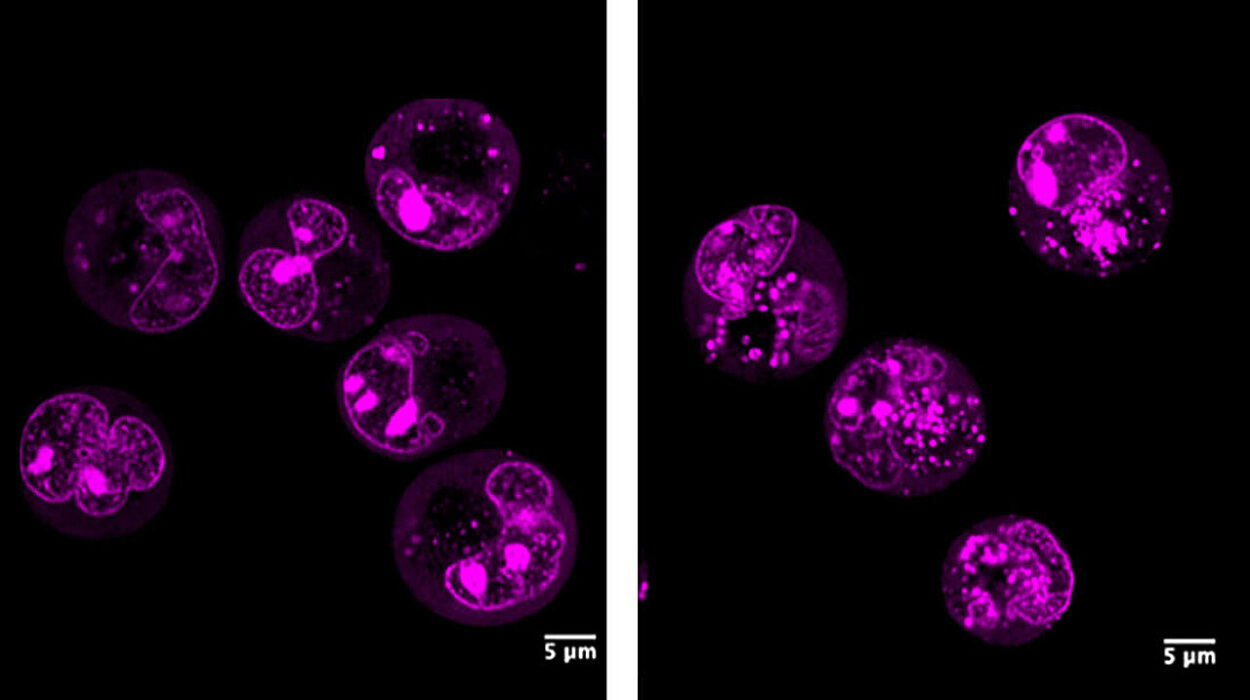
Every cell in your body—each a tenth of a millimeter across—is a bustling city of molecules. Proteins build, move, signal, and defend, orchestrating the ceaseless rhythm of life. Yet beneath all this motion lies a quiet but powerful force: pH, the measure of acidity inside the cell.
Though invisible, pH determines nearly everything that happens within us. It governs how proteins fold, how cells move, and even how they divide. When that delicate balance shifts, the consequences can be devastating—fueling the growth of cancer or the slow damage of neurodegenerative diseases like Alzheimer’s and Huntington’s.
Understanding how cells sense and respond to these tiny pH shifts has long been one of biology’s hardest puzzles. But a groundbreaking study from the University of Notre Dame has now brought scientists closer to solving it.
A New Way to See the Invisible
In research published in Science Signaling, a team led by biochemist Katharine White unveiled a computational method that can scan hundreds of proteins in just days to identify which ones respond to pH changes. This could revolutionize how scientists understand disease at the molecular level—and how they design drugs to treat it.
“Before even picking up a pipette or running a single experiment, we can now predict which proteins are sensitive to pH changes,” said White. “It’s like finding the needle in the haystack before you even start looking.”
Until now, identifying pH-sensitive proteins was painstaking work. Scientists had to study each protein one at a time, slowly testing how it behaved under different pH conditions. Since 1993, only a handful of pH-sensitive proteins were fully characterized, even though researchers suspected that many more existed. Out of thousands of cellular proteins, only about 70 had been confirmed as pH-sensitive—and the exact mechanisms of fewer than 20 were understood.
White’s new computational pipeline changes that.
Mapping the Chemistry of Life
The team’s innovation lies in a powerful algorithm that predicts which parts of a protein are most likely to respond to pH changes. The researchers fed the system structural data from the RCSB Protein Data Bank, a massive global repository that holds the 3D structures of proteins and other biomolecules.
The program then used pKa values—numbers that describe how easily a molecule gains or loses a proton—to estimate how each amino acid’s charge would change across different pH levels. These subtle shifts in electric charge can dramatically reshape how proteins behave.
“Opposite charges attract, and like charges repel,” explained White. “When one amino acid flips its charge, it can set off a cascade of charge-flipping across the entire protein, completely changing its shape and function.”
By mapping these interactions, the program could identify which proteins—and even which exact sites within them—were sensitive to the cellular environment’s acidity. This not only saves years of experimental trial and error but opens the door to studying previously inaccessible biological processes.
Revealing a Hidden Mechanism in Key Proteins
To test their pipeline, the team focused on proteins containing a structure known as the SH2 domain, which acts like a molecular docking site, allowing proteins to link together and relay messages that control growth, division, and immune responses.
Using their model, the researchers predicted that SH2-containing proteins, including SHP2 and c-Src, were sensitive to pH. Laboratory experiments confirmed it.
In SHP2, the team discovered that adding just two protons—tiny particles smaller than atoms—caused the protein’s entire structure to switch from a closed to an open form. This subtle shift effectively turned the protein “on” or “off.”
“This kind of regulation is called allostery,” said White. “It happens away from the protein’s active site but has a huge impact on how the protein functions. It’s incredibly hard to identify, but our approach can now pinpoint it computationally.”
The finding was more than a scientific curiosity—it revealed a molecular mechanism that had eluded researchers for nearly two decades.
How pH Links to Cancer and Disease
One of the study’s most compelling discoveries involved c-Src, an enzyme long known to drive the uncontrolled growth of cancer cells. The Notre Dame team found that in healthy cells, c-Src’s activity depends strongly on pH: it’s most active when the environment is slightly acidic and less active when it becomes more basic.
However, in many cancers, mutations occur right at the pH-sensitive sites identified by the new computational pipeline. These mutations make c-Src insensitive to pH changes—locking it into a constantly active state that fuels relentless cell division and tumor growth.
“Once we knew exactly where the regulation happens, we could see how the mutation breaks it,” said lead author Papa Kobina Van Dyck, who helped design and test the model. “It’s like we condensed 25 years of work into a few weeks.”
This discovery could have enormous implications for drug development. If scientists can design molecules that mimic the natural pH regulation of c-Src, they might be able to switch the enzyme back to its healthy state—shutting down cancer growth without harming normal cells.
Predicting the Future of Precision Medicine
The beauty of White’s computational approach lies in its scalability. What once took years to test can now be done in days, across hundreds of proteins. It’s a leap forward that could transform both basic research and medical treatment.
Daniel DiMaio, deputy director of the Yale Cancer Center, praised the study, noting that “the interior pH of a cell is an important but largely ignored feature because of how difficult it is to measure and manipulate.” With White’s method, he added, researchers finally have “a viable path to analyze the role of intracellular pH,” one that could reveal countless other pH-sensitive processes.
The possibilities extend beyond cancer. pH plays a crucial role in brain function, metabolism, and immune response. Abnormal pH levels are associated with conditions ranging from Alzheimer’s disease and Huntington’s disease to diabetes, autoimmune disorders, and even traumatic brain injury.
By mapping how proteins respond to acidity, scientists can better understand how these diseases begin—and how to stop them.
A New Era for Biochemistry
The research represents a new kind of science: where computation, biology, and medicine converge to decode the living world at its most fundamental level. It also shows how technology can accelerate discovery in ways that were unimaginable just a decade ago.
“Understanding how cells sense their own pH isn’t just about chemistry—it’s about life itself,” White reflected. “If we can learn to read that language, we can learn how to correct what goes wrong in disease.”
In the years to come, her team’s pipeline could help build a comprehensive map of the body’s pH-sensitive proteins—a kind of molecular atlas showing where the delicate balance of acidity keeps us alive, and where its disruption leads to illness.
The Quiet Power of Protons
Every heartbeat, every memory, every movement begins with the flow of invisible particles—protons—moving through the intricate machinery of cells. Their concentration may seem trivial, but it holds the power to shape health, disease, and life itself.
By decoding how these protons interact with proteins, scientists are not just solving one of biology’s long-standing mysteries—they are uncovering a universal principle that could redefine how we treat disease.
Tiny changes, after all, can make all the difference
More information: Papa Kobina Van Dyck et al, Ionizable networks mediate pH-dependent allostery in the SH2 domain–containing signaling proteins SHP2 and SRC, Science Signaling (2025). DOI: 10.1126/scisignal.adt3018