We inherit more than just eye color or height from our parents. Woven into our DNA is a biological story—some chapters are bold and legible, while others are written in a cryptic code scientists are only beginning to translate. For decades, researchers have understood that genes like BRCA1 and TP53 sharply increase cancer risk. But the vast majority of genetic changes we carry—especially those linked to diseases like cancer—fall into a murky category known as variants of uncertain significance, or VUS. They’re biological whispers rather than shouts. They may do nothing at all—or they may be the start of a deadly chain reaction.
Now, in a groundbreaking study published in The American Journal of Human Genetics, researchers from the University of Michigan Medical School are offering powerful new tools to decode those whispers—focusing on a gene called MUTYH, a lesser-known but critically important player in the prevention of colon cancer.
The MUTYH Puzzle: A Common but Clouded Risk
Unlike genes that cause disease outright, MUTYH plays a subtler, but no less essential role: it helps repair damaged DNA. That job is especially vital in fast-dividing tissues like the colon, where DNA replication mistakes can pile up quickly. When MUTYH doesn’t function properly, it can lead to the formation of polyps—abnormal tissue growths that, over time, can develop into colorectal cancer.
What makes MUTYH particularly tricky is that up to 1 in 50 people in the U.S. carry risk variants in this gene. But not all mutations are created equal. Some mutations, known as nonsense variants, shut down the gene entirely—like tearing a page out of a novel. Others, called synonymous variants, are silent and have no effect. But in between lies a spectrum of missense variants—subtle changes that alter one letter in the genetic code, sometimes changing the function of the resulting protein in ways that aren’t easily predictable.
Until now, clinicians have had few tools to determine which MUTYH variants were actually harmful, leaving patients and doctors alike in a state of uncertainty.
Lighting Up the Shadows with Science
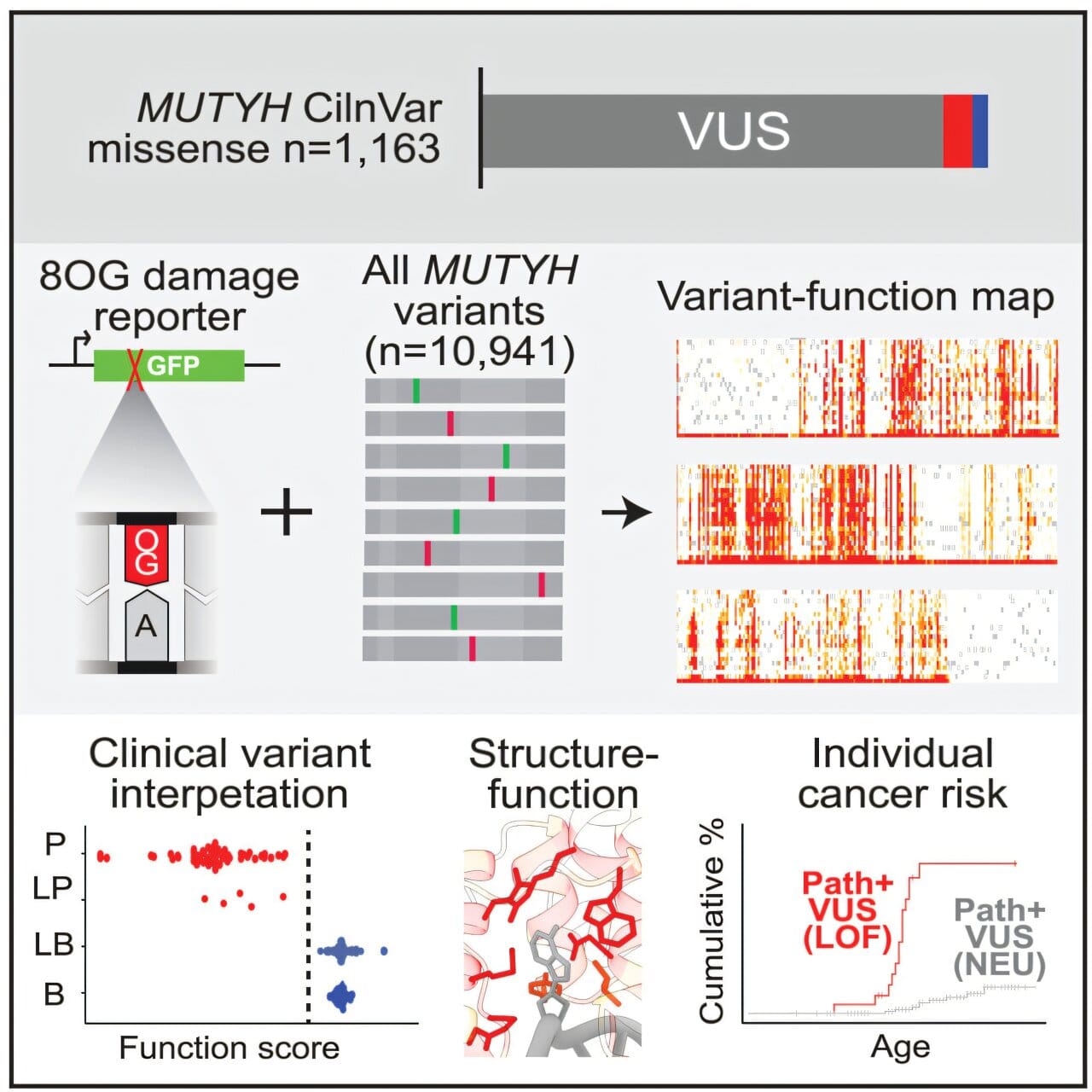
Led by Jacob Kitzman, an associate professor of human genetics, the U-M research team approached this genetic fog not with a flashlight, but with a floodlight. Instead of looking at one mutation at a time—an approach both slow and expensive—they developed a high-throughput method to examine every possible variant of the MUTYH gene, all at once. The result was a mutational library of 10,941 unique versions of the gene.
To test each variant’s impact, the team used a brilliant bit of biotechnology: a DNA-repair reporter system that acts as a fluorescent sensor. Cells containing a functional MUTYH variant glowed green, signaling successful repair of oxidative DNA damage. Cells harboring defective variants remained dim—signaling a breakdown in the gene’s essential repair function.
“It’s like watching a vast field of streetlights,” said Kitzman. “The ones that stay dark tell us where the electrical wiring—in this case, the genetic code—has failed.”
This visual, quantifiable data allowed researchers to sort thousands of variants into two broad categories: functional and non-functional. But even more compelling was the discovery of variants that didn’t fit neatly into either group.
A Gradient of Genetic Risk
Among the 10,000+ tested variants, many missense mutations created a gradient of effects. Some barely affected the gene’s function at all. Others impaired it just enough to increase cancer risk subtly over time. One particular variant fell into this intermediate zone and matched a known clinical form of the disease that causes later-onset, milder polyp formation.
This spectrum of damage is critical because it suggests that genetic risk isn’t binary—it’s a continuum. And with colon cancer, which is often preventable through early detection, knowing where a patient falls on that spectrum could be the difference between life and death.
“This gives us a much more detailed risk map,” Kitzman explained. “For people carrying these variants, we can now begin to say whether their gene is working 100%, 50%, or not at all. That knowledge can directly inform screening and prevention strategies.”
Ground Truth: Comparing with Clinical Data
To validate their results, the researchers cross-referenced their data with the ClinVar database, maintained by the National Center for Biotechnology Information. ClinVar houses thousands of variants collected from genetic testing, along with interpretations from clinical labs regarding their pathogenicity.
The match was striking. Variants known to be harmful lit up as non-functional in Kitzman’s assay. Those classified as benign passed the repair test. And in cases where ClinVar had little or no information—true “variants of uncertain significance”—this new data provides the first solid ground for interpretation.
“In several cases, our functional data perfectly matched the real-world clinical outcomes,” Kitzman said. “That’s a very strong sign that this approach isn’t just academic—it’s actionable.”
The Promise of Personalized Prevention
With the cost of DNA sequencing plummeting, genetic testing is becoming increasingly common. Yet for most people, receiving a genetic test report is like being handed a foreign manuscript: the letters are clear, but the meaning is not.
“People are getting tested more often,” said Kitzman. “But we’re still catching up on how to interpret what we see. We can read the letters in the book, but we don’t know how to turn them into sentences—or what those sentences mean for someone’s health.”
Studies like this are beginning to bridge that gap. For those with a family history of colon cancer, or with troubling but ambiguous test results, the ability to pinpoint whether their MUTYH variant is functional could lead to more personalized—and more effective—preventive care. It might mean earlier colonoscopies, more frequent screenings, or lifestyle changes that help offset genetic risk.
In time, such data could even become part of standard clinical decision-making, transforming vague genetic warnings into clear guidance.
Why Basic Science Still Matters
The study is more than a technological feat—it’s a compelling argument for continued investment in basic biomedical research. Creating a mutational library, developing fluorescent reporter systems, analyzing thousands of variants—these are long-term efforts with no immediate commercial payoff. But they yield priceless benefits.
“This is a case where foundational science connects directly to saving lives,” said Kitzman. “Understanding how genes work isn’t just for textbooks. It can lead to better diagnostics, more accurate risk assessments, and smarter prevention.”
As medicine moves into the era of precision genomics, tools like the one Kitzman’s team has developed will become essential. They don’t just classify risk—they help translate the silent language of DNA into information people can use.
A Brighter Future, One Variant at a Time
Colon cancer remains one of the leading causes of cancer death in the United States, yet it’s also one of the most preventable—if caught early. By unlocking the secrets hidden in genes like MUTYH, researchers are giving patients and physicians a new weapon in the fight: clarity.
It’s a reminder that our genes are not destiny, but dialogue. With the right tools, we can finally start to understand what they’re telling us—and act accordingly.
Kitzman puts it best: “We don’t just want to read the genome. We want to understand it. Because understanding is the first step toward healing.”
More information: Shelby L. Hemker et al, Saturation mapping of MUTYH variant effects using DNA repair reporters, The American Journal of Human Genetics (2025). DOI: 10.1016/j.ajhg.2025.07.005