For centuries, aging has been humanity’s most universal yet mysterious journey—a silent ticking clock that gradually alters our bodies from the inside out. Wrinkles, slower reflexes, and fading stamina are its visible signs, but deep within our tissues, the true story of aging has remained hidden. Now, scientists have unveiled a groundbreaking “proteomic atlas” that charts how aging unfolds across multiple human organs, revealing biological clocks and molecular signals that may drive the entire process.
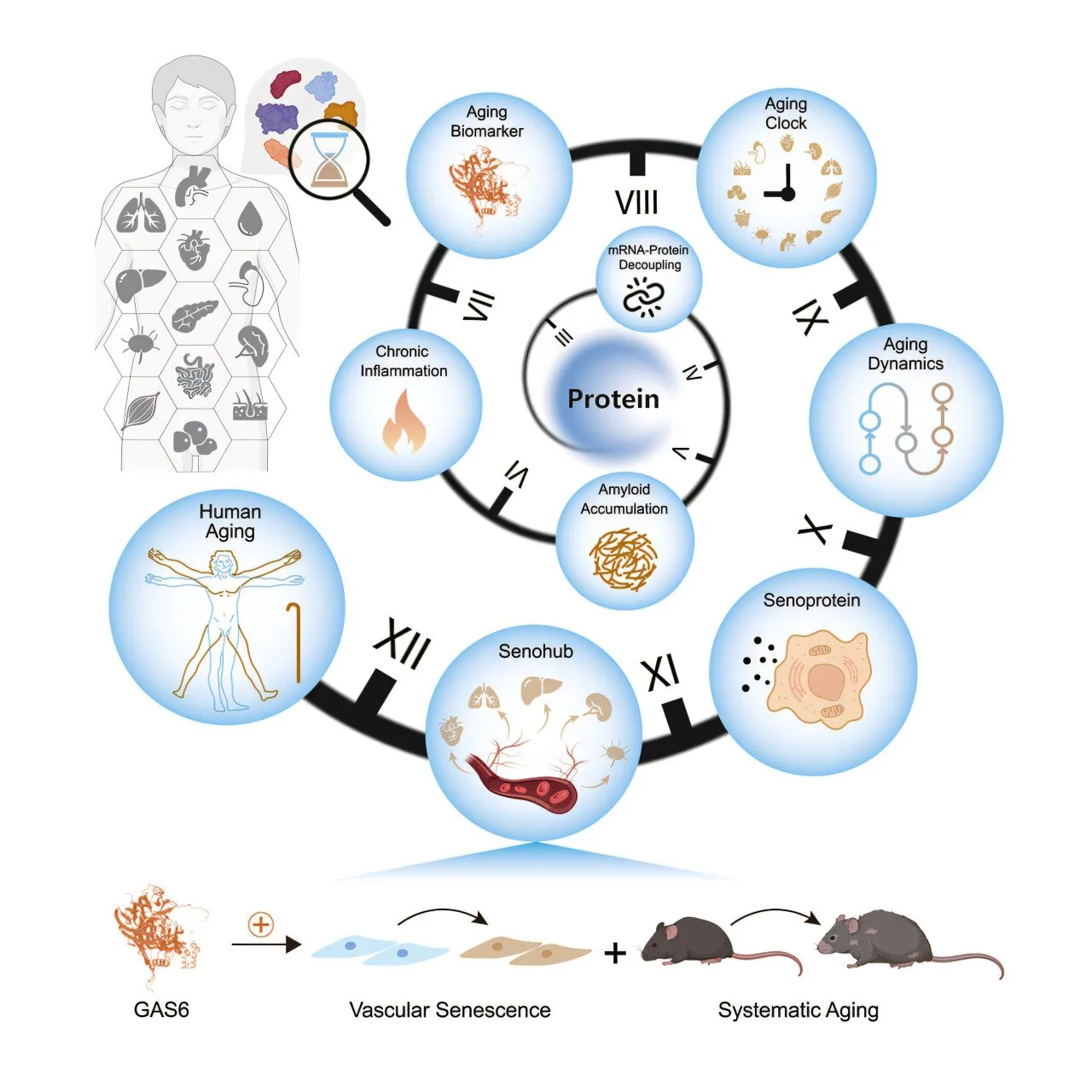
Led by a multi-institutional team spearheaded by the Chinese Academy of Sciences, this landmark study, published in Cell, spans five decades of human life, capturing molecular changes across 13 major organs. Using high-resolution mass spectrometry, researchers measured more than 12,700 proteins from 516 tissue samples and blood plasma, piecing together how proteins—our bodies’ essential building blocks—change with age. The result is the most detailed map of human aging to date, and it sheds light on why different organs age at different rates, why chronic diseases develop, and how certain proteins may accelerate systemic decline.
Beyond DNA: The Protein Landscape of Aging
Previous attempts to measure biological age have largely relied on DNA methylation profiles or circulating plasma proteins. While useful, these approaches overlook what happens at the level of the proteome—the full set of proteins that orchestrate cellular function. Proteins carry out nearly every task in our bodies: they transport oxygen, repair DNA, fold into enzymes, and act as molecular signals between cells. Understanding their life cycle is critical for understanding aging itself.
This new study reveals that protein quality control—the mechanisms that synthesize, fold, and degrade proteins—deteriorates with age, but not uniformly. Different organs show unique trajectories. For example, muscle, spleen, and lymph nodes experience a pronounced breakdown in the coordination between RNA and protein expression. This means that even when a gene remains active, the corresponding protein may fail to be produced correctly, leading to cellular malfunction.
In nearly every tissue studied, the researchers found a loss of correlation between transcriptome (mRNA levels) and proteome (protein levels), signaling a fundamental disruption in biological communication. The machinery responsible for protein folding and synthesis, including ribosomes and chaperonins, also declined, potentially paving the way for dysfunctional proteins to accumulate.
The Amyloid Storm and Cellular Inflammation
Among the most striking findings was the accumulation of amyloid proteins—misfolded protein aggregates linked to diseases like Alzheimer’s—as well as immunoglobulins and complement factors. Together, these form what the authors describe as an “amyloid-immunoglobulin-complement axis,” a vicious cycle in which misfolded proteins trigger immune activation and inflammation.
This axis was not limited to the brain; it appeared in multiple tissues, suggesting that amyloid-related processes might drive systemic aging far beyond neurodegenerative disease. One protein in particular, serum amyloid P-component (SAP), increased steadily in multiple organs. Functional tests showed that SAP could directly induce cellular senescence and inflammation, turning healthy vascular cells into aged, dysfunctional ones.
Biological Age Isn’t the Same Everywhere
To quantify aging in each organ, researchers built tissue-specific “proteomic clocks” using advanced machine learning models. These clocks could predict the biological age of an organ with up to 95% accuracy. Intriguingly, the aging curves were not synchronized. The aorta—the largest artery in the body—showed sustained and early aging signals, while other tissues aged more slowly.
Between ages 45 and 55, most organs exhibited an inflection point—a midlife molecular tipping point where protein degradation pathways faltered more dramatically. This suggests that interventions targeting this critical window might slow or even partially reverse organ decline before it cascades into chronic disease.
A recurring protein in these aging clocks was TIMP3, an inhibitor of enzymes that remodel tissues. Its presence across nine distinct clocks hints at a universal role in vascular stiffening and extracellular matrix changes that affect multiple organs simultaneously.
The Body’s Aging Messengers: Secreted Proteins
Perhaps the most profound discovery was the identification of proteins secreted by aged tissues that appear to spread senescence signals throughout the body. These blood-borne factors, including chemokines and adhesion molecules, act as molecular messengers between organs.
One such protein, GAS6, was elevated in aging plasma and in the aorta. Experiments showed that GAS6 could induce senescence markers, provoke inflammation, impair blood vessel growth, and increase immune cell adhesion. When researchers injected GAS6 into middle-aged mice, the animals developed vascular dysfunction, tissue inflammation, and physical decline, mimicking features of human aging.
Another secreted protein, GPNMB, had similarly profound effects. In both human cell cultures and live mice, GPNMB promoted vascular inflammation and tissue degeneration, leading to impaired motor performance. These findings reveal a previously hidden layer of inter-organ communication—an aging “signal network” that may amplify decline beyond individual tissues.
The study’s authors coined the term “senohub” to describe organs like the aorta that appear to act as central initiators of systemic aging, sending out distress signals that gradually pull the rest of the body into senescence.
Why This Matters for Chronic Diseases
Chronic illnesses such as cardiovascular disease, diabetes, and neurodegeneration often emerge in mid-to-late life. This research suggests a possible molecular explanation: as protein quality control breaks down, misfolded proteins accumulate, inflammation spreads, and tissues begin to biologically age at different speeds. The vascular system, in particular, seems to play a critical role as both an early sensor and driver of aging, potentially explaining why cardiovascular health is so strongly linked to overall longevity.
Understanding these pathways opens new possibilities for therapeutic interventions. If scientists can block harmful proteins like GAS6 or GPNMB, or bolster protein maintenance systems that falter with age, it might be possible to slow the biological aging of multiple organs simultaneously, delaying the onset of chronic diseases.
A Map for the Future
Beyond its immediate findings, this proteomic atlas represents a foundational resource for future research. By cataloging how more than 12,700 proteins change over a 50-year lifespan, it gives scientists a detailed molecular roadmap of human aging. This roadmap will allow researchers to pinpoint early markers of age-related decline, design drugs that restore protein balance, and test anti-aging therapies with unprecedented precision.
The work also highlights the importance of studying aging as a multi-organ phenomenon rather than focusing solely on blood markers or genetic changes. It reminds us that aging is not just a passive countdown but an active, dynamic process shaped by interwoven molecular dialogues within and between tissues.
Toward Healthy Aging
While the passage of time is inevitable, this research hints that the rate and nature of biological aging might not be fixed. Just as a failing clock can be repaired by addressing its broken gears, our organs may one day be rejuvenated by rebalancing their molecular machinery. The proteomic atlas brings us closer to that future, where aging is not merely endured but understood and, perhaps, better managed.
In the quiet language of proteins—folding, misfolding, signaling, and secreting—the body writes its own story of aging. For the first time, scientists have learned to read that story in detail, unlocking a new chapter in our quest to understand and one day master the biology of time itself.
Reference: Yingjie Ding et al, Comprehensive human proteome profiles across a 50-year lifespan reveal aging trajectories and signatures, Cell (2025). DOI: 10.1016/j.cell.2025.06.047