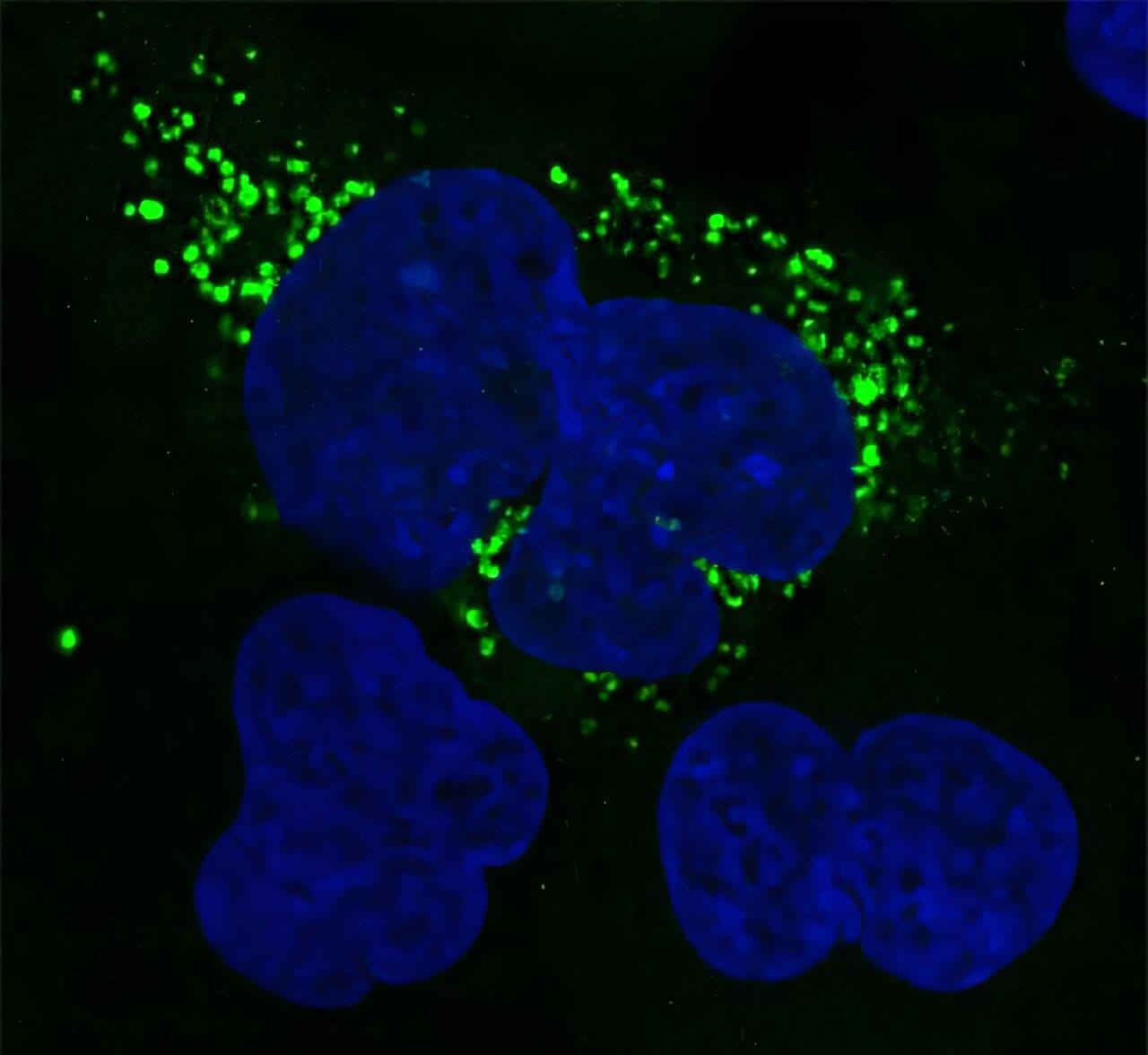
In the grand orchestra of life, proteins play the symphony. These molecular machines fold, twist, and dance into nearly every biological role—structural support, catalyzing reactions, regulating genes. Yet for decades, scientists have been blind to a hidden ensemble—microproteins, tiny proteins composed of fewer than 150 amino acids that lurk in what was once dismissed as the dark matter of our DNA.
Now, a groundbreaking study from the Salk Institute is helping to bring these elusive microproteins into the spotlight. Armed with a new machine learning tool called ShortStop, researchers are sifting through vast genomic databases to locate, classify, and prioritize these once-overlooked molecular players. Their findings suggest that microproteins may hold crucial clues to diseases like cancer and Alzheimer’s, ushering in a new chapter in molecular biology.
The Undiscovered Country of the Genome
Only about 1% of the human genome codes for what we traditionally define as proteins—those large, well-characterized chains that run the cellular machinery. The rest, once infamously labeled “junk DNA,” was long believed to be evolutionary leftovers, with little to no functional role. But science has a way of humbling assumptions.
Hidden within this so-called junk lie small open reading frames (smORFs), tiny stretches of DNA with the potential to code for microproteins. These microproteins have been obscured not just by scientific dogma but by technical limitations. Because they’re so small, they’re difficult to detect with standard tools like mass spectrometry or genome annotation software, which tend to overlook sequences below a certain threshold.
Imagine scanning a crowd for giants while ignoring the children. That’s how science treated microproteins for decades—until now.
Enter ShortStop: A Machine Learning Game-Changer
Developed by the lab of Professor Alan Saghatelian at the Salk Institute, ShortStop is a clever and efficient tool trained to find the needles in the genomic haystack. It’s not just a data scraper—it’s a prioritization engine.
ShortStop works by comparing real smORFs from genetic datasets against a set of artificial “decoy” smORFs—sequences generated by computer algorithms that mimic nonfunctional genetic noise. This two-class training model teaches the algorithm to distinguish potential functional microproteins from biological static.
The result is an elegant leap forward in genomic discovery. Instead of spending weeks testing hundreds or thousands of sequences, researchers can now focus on the few that stand out as most promising. In essence, ShortStop does the hard work of triage, vastly reducing wasted time and laboratory resources.
What Makes Microproteins So Mysterious?
Microproteins are small—often no more than a few dozen amino acids long—and it’s exactly this modest size that has made them invisible to traditional biochemical techniques. For comparison, average proteins like insulin contain over 50 amino acids, and some stretch into the thousands.
These microproteins may seem insignificant at first glance. But biological importance doesn’t scale with size. Recent studies have implicated microproteins in everything from muscle development to stress responses in cells. Many are evolutionarily conserved, which hints that natural selection preserved them for a reason.
Some microproteins act like switches, binding to larger proteins and altering their activity. Others may fine-tune gene expression or act as signals within cells. They are molecular whispers in a symphony of shouts—and they may be just as essential.
A Hidden Signal in Lung Cancer
ShortStop isn’t just a theoretical advance—it’s already bearing fruit. In their recent study, the Salk team applied the tool to a publicly available dataset of lung cancer tissue, comparing genetic information from tumor cells and nearby normal cells. Among the 210 newly identified microprotein candidates, one stood out: a microprotein that was significantly upregulated in cancerous tissue.
This specific microprotein had never been seen before, and yet, it was more active in tumors than in healthy lung tissue. That kind of pattern is a red flag—and a tantalizing opportunity. The protein might serve as a biomarker, helping doctors detect lung cancer earlier, or it could even represent a new therapeutic target, a molecular vulnerability that future drugs could exploit.
This single example underscores the enormous untapped potential of re-analyzing existing data. With ShortStop, genetic archives become treasure chests.
Rewriting the Rules of Molecular Biology
What makes this discovery particularly poignant is how it challenges long-held assumptions. Molecular biology, since its mid-20th-century golden age, has largely focused on big, obvious targets—genes with clear functions, proteins with prominent roles. But science evolves, and the frontier is always shifting.
Microproteins force us to rethink what’s biologically meaningful. For decades, smORFs were ignored because they didn’t conform to existing models. Now, with tools like ShortStop, we’re learning that biology doesn’t care about our models—it follows its own code, often subtle, always complex.
Professor Saghatelian puts it succinctly: “Most of the proteins in our body are well known, but recent discoveries suggest we’ve been missing thousands of small, hidden proteins.” These aren’t rare mutations or fringe curiosities. They may be integral parts of our biology, waiting to be understood.
Implications for Health and Disease
The potential impact of microproteins touches nearly every corner of medicine. Diseases like Alzheimer’s, diabetes, and cancer often involve complex regulatory networks within cells. Microproteins could be the missing pieces in these puzzles—tiny regulators that explain why treatments fail, why symptoms differ, why cells behave erratically.
Moreover, microproteins could help personalize medicine. If some people express certain microproteins and others don’t, that variability could explain differences in drug response or disease susceptibility. ShortStop allows scientists to explore those differences quickly, at scale, and with unprecedented precision.
Brendan Miller, the study’s lead author and a postdoctoral researcher in Saghatelian’s lab, highlights another practical advantage: “ShortStop works with common data types, like RNA sequencing datasets, which many labs already use.” That means labs worldwide can begin their own microprotein hunts, accelerating discovery far beyond the walls of the Salk Institute.
From Junk DNA to Genetic Goldmine
Perhaps the most poetic aspect of this discovery is what it reveals about the genome itself. For years, we were taught that most of our DNA was inert—vestiges of evolution, clutter without purpose. But science thrives on challenging assumptions, and the story of microproteins reminds us that what we call “junk” may simply be what we don’t yet understand.
ShortStop is helping to transform our view of the genome from a linear instruction manual into a multilayered, dynamic system full of hidden messages. These microproteins may be the Morse code of the genome—brief, often cryptic, but packed with meaning.
And the search is just beginning. The Salk team believes that microproteins may number in the thousands. Each one has the potential to be a switch, a signal, or a sentinel. Each one could unlock a new chapter in our understanding of human biology.
A Future Fueled by the Smallest Discoveries
As the scientific community continues to push the boundaries of what we know, the discovery and characterization of microproteins stand as a testament to the power of looking where no one thought to look.
Tools like ShortStop represent more than just algorithms—they are amplifiers of human curiosity, engines of exploration that make the invisible visible. They allow us to see not just more data, but new kinds of data, buried in places we once ignored.
In the quiet stretches of DNA once thought silent, a chorus of microproteins may be singing the hidden songs of life. With ShortStop, we’ve begun to hear their first notes.
And they are profound.
More information: ShortStop: A machine learning framework for microprotein discovery, BMC Methods (2025). DOI: 10.1186/s44330-025-00037-4