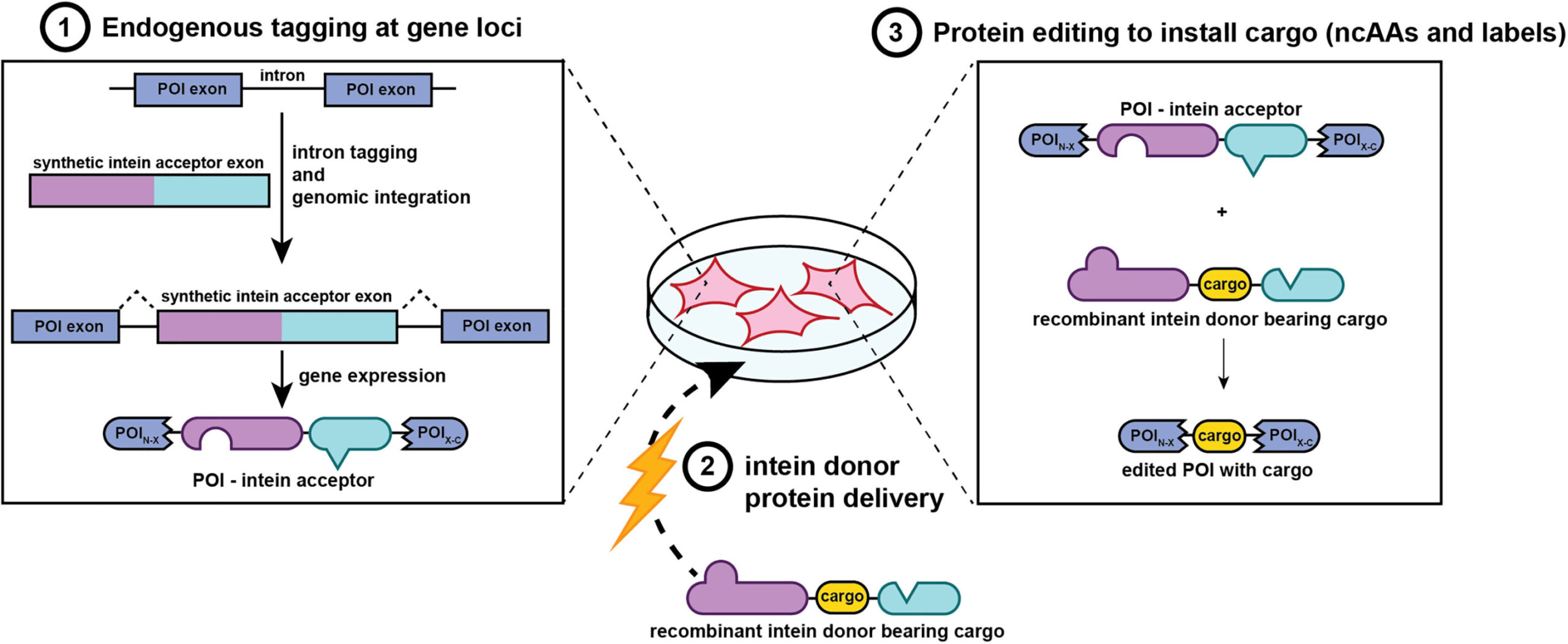
In a quiet lab nestled within the University of Pennsylvania, scientists have unlocked a powerful new way to watch life unfold from the inside. With precision and speed once thought unattainable, researchers have developed a groundbreaking technique that allows proteins—the microscopic engines of biology—to be edited, labeled, and tracked directly within living mammalian cells. This new tool opens a window into the secret inner workings of cells, providing scientists with an unprecedented ability to study biology in its most natural state: alive, intact, and in motion.
Published in Science under the title “Intracellular protein editing enables incorporation of noncanonical residues in endogenous proteins”, this innovative research not only solves a long-standing challenge in cell biology but also charts a bold new path for understanding disease, designing better drugs, and decoding the language of life itself.
Why Study Proteins in Living Cells?
Proteins are the artisans and architects of the cell. From building cellular scaffolds to catalyzing chemical reactions and regulating gene expression, nearly every aspect of life hinges on the work of proteins. But understanding how proteins do their jobs—when they turn on, where they go, and what other proteins they interact with—requires seeing them in their natural habitat: the living cell.
Yet studying proteins in real-time has always been a delicate affair. Traditional tools like fluorescent proteins and epitope tags are often bulky. Like trying to study a dancer by watching them wear a backpack, these tags can interfere with the protein’s natural rhythm—altering its movement, changing its structure, or even sending it to the wrong place inside the cell. Small-molecule labels, while less intrusive, are notoriously difficult to apply inside live cells without causing damage or disruption. Antibodies, though highly specific, are generally incompatible with the dynamic, ever-changing environment of living tissue.
Moreover, many existing protein labeling methods are slow or lack the temporal resolution needed to capture fleeting events—like signaling cascades that occur in seconds, or adaptive responses to stress that unfold in minutes. The field has long needed a technology that combines precision, speed, and subtlety, working quietly and efficiently within the living cell without disrupting the act it seeks to observe.
A New Tool for Editing Life at the Molecular Level
The University of Pennsylvania team has now met this unmet need head-on with a protein-editing system that is nothing short of molecular surgery. This method enables researchers to edit protein sequences inside living mammalian cells by inserting chemical tags, artificial amino acids, or peptide segments into either native (endogenous) or engineered (exogenous) proteins.
At the heart of this method are two finely tuned molecular tools—engineered splicing elements that function like programmable editing switches. When a donor protein carrying the desired modification is introduced into the cell, these editing switches activate, directing the insertion of the new segment precisely where it’s needed. Whether the goal is to add a fluorescent label, insert a therapeutic peptide, or restore a mutated region to its original sequence, this system provides a clean, seamless method for modifying the protein on demand.
Even more impressive is the finesse of this operation. Once the insertion is complete, the editing tools themselves are removed from the final product, leaving behind no molecular fingerprints or scars. The protein remains fully functional, indistinguishable from its original form except for the intentional edit—just as if nature had placed it there herself.
Precision in Minutes: A Leap in Speed and Specificity
In one of the most stunning revelations of the study, the editing process unfolds within just 10 minutes of delivering the donor protein into the cell. This rapid timeline marks a dramatic improvement over traditional tagging methods, which often require hours or even days to produce results. Speed matters: in the fast-paced world of cellular signaling, many important biological decisions are made in seconds. This method gives researchers a front-row seat to those decisions, right as they happen.
To test the technique, the scientists used model proteins including β-actin, a core component of the cytoskeleton, and calnexin, a protein critical to the endoplasmic reticulum. Using a combination of immunoblotting, mass spectrometry, and microscopy, they confirmed that the inserted sequences were present, the proteins remained active, and cellular localization was preserved. The edited proteins continued to perform their essential functions, proving that the method was not only fast and precise but also biologically respectful.
Editing Essential Proteins Without Disruption
In perhaps the most convincing validation of the system’s power, the researchers moved on to editing proteins with critical cellular roles. Among these were c-Myc, a well-known transcription factor involved in cell growth and cancer, and Chk1, a kinase that regulates the DNA damage response.
These proteins are not forgiving of interference. Yet even here, the editing method showed remarkable grace. Specific segments were temporarily replaced with identifiable tags—allowing scientists to track and study them in real time—before being restored to their original sequences. Despite the editing, both c-Myc and Chk1 retained full functionality, as confirmed by normal phosphorylation behavior and intact DNA-binding capacity.
This ability to edit without breaking is crucial. It means that scientists can now study even the most sensitive or essential proteins without fear of corrupting the data or disrupting cellular integrity.
Fluorescent Tracers and Synthetic Handles
The applications of this technology extend far beyond basic tagging. By incorporating noncanonical amino acids—amino acids not found in nature but designed in the lab—researchers can now introduce synthetic chemical handles into proteins. These handles act like docking stations for other molecules, allowing scientists to attach fluorophores, biotin, or even drug-like compounds in a controlled, modular fashion.
In experiments using β-actin and calnexin, donor proteins carrying synthetic handles were first labeled in vitro with desired tags and then introduced into mammalian cells. Once inside, these modified proteins were successfully spliced into their targets. The edited proteins localized to their expected cellular compartments and emitted strong fluorescent signals that could be observed under confocal microscopy. Moreover, the modified proteins could be extracted using affinity purification techniques, opening the door to high-resolution studies of protein-protein interactions and post-translational modifications.
Versatile Delivery: Electroporation and Lipid Nanoparticles
No molecular tool is complete without an efficient delivery system. To bring the editing machinery into the cell, researchers tested both electroporation and lipid nanoparticle (LNP) methods. Electroporation—using an electric field to open pores in the cell membrane—proved effective but can sometimes damage sensitive cells.
Lipid nanoparticles offered a gentler alternative. Especially when the donor proteins were engineered with negatively charged tails, LNPs delivered the editing components with high efficiency. This formulation could be particularly useful in therapeutic applications, where minimizing cellular stress is a priority.
The ability to edit proteins without disrupting the cell’s natural behavior—combined with flexible delivery strategies—makes this technique suitable for a wide range of cell types and experimental conditions.
Clear Signals, Low Background, Real-Time Insight
In live-cell imaging, one of the biggest challenges is background noise—signals from non-specific labeling, misfolded proteins, or degraded components that muddy the picture. Here, too, the new editing method shines.
Fluorescent signals from properly edited proteins matched the known localization of the targets. Calnexin appeared neatly within the endoplasmic reticulum; β-actin assembled in the cytoskeleton, forming beautiful filaments stretching across the cell. In contrast, signals from unedited or incorrectly delivered proteins faded quickly, indicating that background noise was minimal and easily distinguishable from genuine labeling.
This clarity enables researchers to follow protein dynamics over time—watching as molecules migrate, interact, or respond to external cues in real time.
A Universe of Possibilities: From Cell Biology to Drug Design
The implications of this technology are vast. For molecular biologists, it offers a new lens through which to observe the living cell—not just in snapshots, but in continuous, dynamic detail. For medical researchers, it provides a tool to study disease-relevant proteins in their native context, potentially revealing vulnerabilities that can be targeted with next-generation therapies.
Imagine tagging cancer-driving proteins in real-time to understand how they interact with tumor-suppressing partners. Picture introducing therapeutic peptides into defective proteins, correcting genetic errors at the protein level without altering DNA. Envision drug developers screening thousands of molecules against live, labeled proteins inside actual cells—not test tubes—producing more accurate, faster, and safer results.
This technology is not just a method. It is a platform—one that bridges the gap between synthetic biology, live-cell imaging, and personalized medicine.
Looking Ahead: The New Language of Life
In many ways, the discovery marks a turning point in cellular biology. For decades, scientists have struggled with the challenge of studying life without disturbing it. This new editing technique, by allowing rapid, precise, and gentle protein modification within live mammalian cells, makes that dream a reality.
It is a tool built not just for understanding how cells function, but for redefining how we interact with them—enabling interventions at the molecular level that are as sophisticated and nuanced as biology itself.
As science continues its voyage into the inner world of the cell, the researchers at the University of Pennsylvania have handed us a compass. One that points not north, but inward—toward the molecular truth of life, and the untapped potential waiting within every protein, every cell, every breath of living matter.
Reference: Jenna N. Beyer et al, Intracellular protein editing enables incorporation of noncanonical residues in endogenous proteins, Science (2025). DOI: 10.1126/science.adr5499. www.science.org/doi/10.1126/science.adr5499