In the hidden corners of biology, where molecules twist and dance in realms far smaller than the eye can see, scientists have made a surprising discovery—one that could rewrite how we fight HIV and perhaps even reimagine our approach to autoimmune diseases. For decades, lipids—those oily, slippery molecules that form the outer membranes of our cells—were seen as off-limits to antibodies. These fatty molecules are vital to life, shared between viruses and healthy human cells alike, and targeting them was considered far too risky. But what if the very lipids we’ve shied away from could become the key to unlocking one of medicine’s greatest challenges?
A team of researchers at Scripps Research has flipped this conventional wisdom on its head. Using sophisticated computer modeling, they’ve shown how a rare class of antibodies known as broadly neutralizing antibodies (bNAbs) use lipids not as obstacles, but as signposts—biological cues that guide them to the HIV virus. Their findings, published in eLife on April 7, 2025, offer a new blueprint for vaccine design, potentially opening a path not just for better HIV therapies, but for innovations in treating autoimmune disorders as well.
“This work gives us a clearer picture of how these antibodies interact with the viral membrane, giving a blueprint we can potentially apply to vaccine design,” says Marco Mravic, an assistant professor at Scripps Research and a senior author of the study.
Lipids: The Unlikely Guideposts of Immunity
Lipids are everywhere. They coat every cell in your body, forming bilayers—double-walled membranes that keep the inside of cells separate from the chaotic soup outside. They’re also present in viruses, which use similar membranes to protect their genetic payloads. This ubiquity has made them poor candidates for immune system targeting. Target a lipid, and you risk targeting yourself.
That’s why scientists long believed antibodies couldn’t safely lock onto lipids without triggering autoimmune havoc. But as it turns out, biology—as always—has a few elegant tricks up its sleeve.
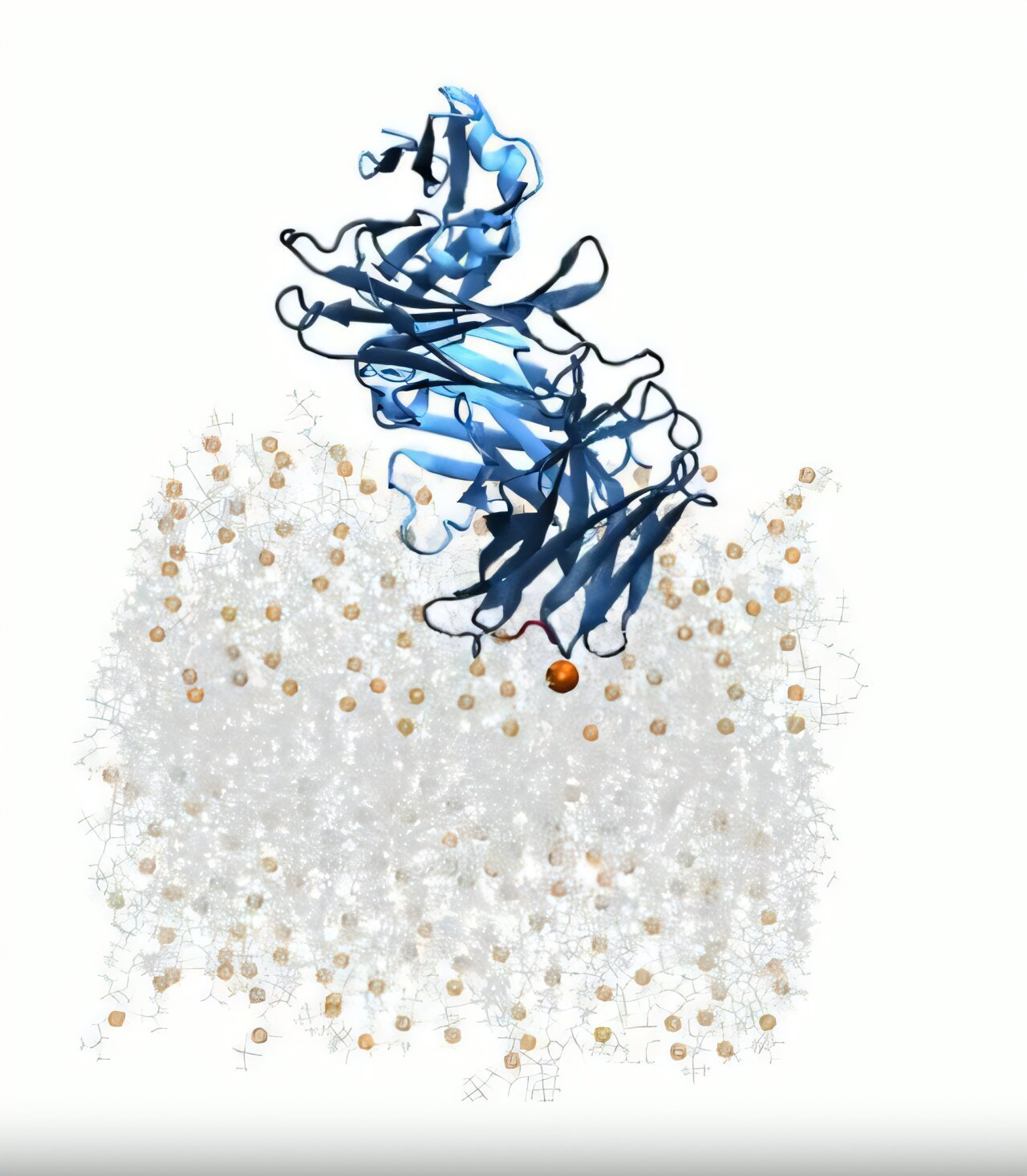
At the center of this new discovery is a region of the HIV virus called the membrane-proximal external region, or MPER. Located just at the surface of the virus’s membrane, this region is tantalizingly exposed—but frustratingly hard to reach. It’s like trying to grab a coin at the bottom of a swimming pool without disturbing the water.
“MPER sits right at the lipid bilayer—or membrane—on the HIV cells,” explains Colleen Maillie, the study’s first author and a joint Ph.D. candidate in the Mravic and Andrew Ward labs. “It’s notoriously difficult to reach because it’s concealed.”
Simulating the Unseeable
The key to reaching this elusive target wasn’t brute force, but nuance—and a leap in technology. Rather than rely solely on traditional experimental methods, which struggle with the complex interplay between proteins and lipids, the Scripps team turned to computational modeling. These multi-scale simulations allowed them to explore interactions at atomic precision, visualizing how antibodies approached, attached, and stabilized at the membrane interface.
What they found was nothing short of revolutionary. Two distinct features of the antibodies stood out: loop regions and framework regions.
Loop regions are the flexible, wavy structures on the tip of the antibody—the part that usually binds to a target. These loops were already known to play a role in recognizing the MPER. But it was the framework regions, once thought of as inert scaffolding, that provided the surprise. These sheet-like structures appeared to form intimate contact with the lipid bilayer, helping orient the antibody and increasing its grip.
“That was really unexpected,” Maillie says. “Framework regions are hardwired into antibodies from birth, and they weren’t thought to play much of a role in targeting. But here, they seem to be guiding the whole interaction.”
This idea lends critical support to a vaccine design strategy pioneered at Scripps Research called germline targeting, developed by Professor William Schief. The concept involves designing immunogens—substances that provoke an immune response—that can stimulate these early framework structures in a way that steers antibody development down a favorable evolutionary path.
Lipid First, Virus Later?
A central debate in HIV immunology has been whether antibodies recognize the virus’s lipid membrane first and then drift toward the protein-rich MPER, or if they latch onto MPER and incidentally encounter the membrane afterward. The Scripps simulations suggest the former may be true. By modeling antibodies interacting with membranes made entirely of lipids, the researchers showed that lipid binding can precede and guide protein binding.
“That’s a major insight,” Maillie notes. “And it’s only possible to see that kind of sequence of events in simulations. In real-world experiments, you can’t isolate time in that way.”
Equally striking was the fidelity of these simulations. The team reproduced molecular interactions with such high accuracy that the models nearly mirrored experimental observations.
“It was really cool to see that these physics-based approaches in our simulations are able to accurately model what we see in nature,” Maillie says. “If we have that kind of accuracy, what else can we determine?”
Walking the Tightrope of Autoimmunity
But there’s a catch. If antibodies can bind to lipids—molecules shared across all cell membranes—what’s to stop them from turning on the body itself?
This is a question that cuts to the heart of autoimmune diseases like lupus, in which the immune system mistakenly targets the body’s own cells. Traditionally, an increase in lipid affinity was seen as a red flag—a potential indicator of autoimmunity. But when the Scripps team modeled antibodies at different stages of their development, they found something surprising.
“As the antibodies matured, their affinity for the lipid component of the virus increased,” Maillie explains. “That was shocking. It suggests that the immune system is tolerating these higher lipid interactions, which we didn’t expect.”
The difference may lie in specificity. Autoimmune antibodies tend to interact loosely and indiscriminately with lipids. The antibodies studied here, however, showed an exquisitely precise recognition of defined lipid-protein structures—a precision that could distinguish friend from foe at the molecular level.
“The structure of the antibody itself holds an encoding principle to recognize lipids,” Maillie says. “That’s something that was really underappreciated before.”
Implications for Vaccines, Autoimmune Therapy, and Synthetic Antibodies
The discovery that lipids can act as a kind of ‘molecular landing pad’ for antibodies could have far-reaching consequences.
In HIV research, vaccine developers are already working to design antigens that can stimulate bNAbs capable of targeting the MPER with both lipid and protein specificity. This dual recognition could be the key to creating vaccines that offer long-lasting protection across different strains of the virus.
But the implications extend well beyond HIV. Understanding how antibodies develop lipid-recognition traits without tipping into autoimmunity could shed light on conditions like lupus, multiple sclerosis, and rheumatoid arthritis—diseases in which the body’s immune system turns against its own membranes.
And then there’s synthetic biology. One of the grand challenges in therapeutic development is designing molecules that can recognize complex targets on the cell membrane, such as ion channels and G-protein-coupled receptors (GPCRs). These membrane proteins are involved in everything from nerve signaling to hormone reception, making them prime targets for drugs—but they’re notoriously hard to reach.
Now, researchers may be able to design synthetic antibodies that mimic the lipid-targeting strategies of bNAbs, expanding the toolbox for drug development.
“Synthetic protein engineers can look at this and say, ‘The immune system tolerates these specific lipid-targeting elements,’” Maillie says. “Maybe other antibodies can start to incorporate those and use them to their advantage.”
A New Lens on Immunity
The study, titled “Ab initio prediction of specific phospholipid complexes and membrane association of HIV-1 MPER antibodies by multi-scale simulations,” may have focused on HIV, but its ripples extend across immunology. It challenges long-held assumptions about what antibodies can safely target. It illustrates how lipids, once viewed as immunological minefields, can be part of the solution. And it demonstrates the power of simulation—not just as a tool for visualizing biology, but as a window into the rules that govern life itself.
As scientists continue to decode the language of lipids, they may find that the cell membrane—long thought of as biology’s inert wallpaper—is actually rich with signals waiting to be understood. In the folds of fat and protein, nature has embedded clues, and we’re only just beginning to read them.
Reference: Colleen A Maillie et al, Ab initio prediction of specific phospholipid complexes and membrane association of HIV-1 MPER antibodies by multi-scale simulations, eLife (2025). DOI: 10.7554/eLife.90139.3